Using Population objects to create biased data#
import mlsim
import pandas as pd
import numpy as np
import seaborn as sns
from collections import namedtuple
Create an all default population
pop = mlsim.bias.Population()
To view the details on this population, we can use the get_parameter_description method.
print(pop.get_parameter_description())
Demographic Parameters
DemParams(Pa=[0.5, 0.5], Pz_a=[[0.5, 0.5], [0.5, 0.5]])
Target Parameters
TargetParams(Py_az=[[[0.95, 0.05], [0.95, 0.05]], [[0.95, 0.05], [0.95, 0.05]]])
Feature Parameters
FeatureParams(distfunc=<function <lambda> at 0x7f367781e700>, theta=[[[[5, 2], [2, 5]], [[5, 2], [2, 5]]], [[[5, 2], [2, 5]], [[5, 2], [2, 5]]]])
Feature Noise Parameters
NoiseParams(noisefunc=<function <lambda> at 0x7f367781f420>, theta=[[[1.0, 1.0], [1.0, 1.0]], [[1.0, 1.0], [1.0, 1.0]]])
The instantiation just assigns values to these parameters. In order to get data, we use the sample method.
help(pop.sample)
Help on method sample in module mlsim.bias.populations:
sample(N, return_as='DataFrame') method of mlsim.bias.populations.Population instance
sample N members of the population, according to its underlying
distribution
Parameters
-----------
N : int
number of samples
return_as : string, 'dataframe'
type to return as, can be pandas 'DataFrame' or IBM AIF360
'structuredDataset'
pop_df1 = pop.sample(100)
pop_df1.head()
| a | z | y | x0 | x1 | |
|---|---|---|---|---|---|
| 0 | 0.0 | 1.0 | 1.0 | 3.182082 | 4.594324 |
| 1 | 1.0 | 1.0 | 1.0 | 3.049347 | 6.543984 |
| 2 | 0.0 | 1.0 | 1.0 | 0.813956 | 7.642841 |
| 3 | 1.0 | 0.0 | 0.0 | 4.386188 | 2.503460 |
| 4 | 0.0 | 0.0 | 0.0 | 2.733758 | 4.678923 |
Changing the type of bias#
Now demo some with various biases to create examples
# create a correlated demographic sampler
label_bias_dem = mlsim.bias.DemographicCorrelated(rho_a=.2,rho_z=[.25,.15])
# instantiate a population with that
pop_label_bias = mlsim.bias.PopulationInstantiated(demographic_sampler=label_bias_dem)
pop_label_bias_df1 = pop_label_bias.sample(100)
pop_label_bias_df1.head()
| a | z | y | x0 | x1 | |
|---|---|---|---|---|---|
| 0 | 0.0 | 0.0 | 0.0 | 5.059749 | 0.462498 |
| 1 | 0.0 | 0.0 | 0.0 | 3.938205 | 3.445628 |
| 2 | 1.0 | 1.0 | 1.0 | 2.576376 | 7.430619 |
| 3 | 0.0 | 0.0 | 0.0 | 3.784182 | 3.497882 |
| 4 | 0.0 | 1.0 | 1.0 | 4.630621 | 5.814778 |
New we’ll create a feature bias where the classes are separable for one group and not for the other.
feature_sample_dist = lambda mu,cov :np.random.multivariate_normal(mu,cov)
per_group_means = [[[1,2,3,4,3,3],[4,6,8,8,10,6]],[[3,2,3,4,4,3],[1,3,4,4,5,3]]]
D =6
shared_cov = [np.eye(D)*.75,.95*np.eye(D)]
feature_bias = mlsim.bias.FeaturePerGroupSharedParamWithinGroup(
feature_sample_dist,per_group_means,shared_cov)
pop_feature_bias = mlsim.bias.PopulationInstantiated(feature_sampler=feature_bias)
pop_feature_bias_df1 = pop_feature_bias.sample(100)
pop_feature_bias_df1.head()
| a | z | y | x0 | x1 | x2 | x3 | x4 | x5 | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | 0.0 | 0.0 | 1.016204 | 0.488367 | 1.198120 | 4.140980 | 2.847464 | 0.083858 |
| 1 | 0.0 | 1.0 | 1.0 | 3.660997 | 4.850933 | 6.043325 | 4.757886 | 12.464587 | 4.044343 |
| 2 | 1.0 | 1.0 | 1.0 | 1.108399 | 3.132314 | 5.397789 | 3.341915 | 4.324666 | 6.274768 |
| 3 | 0.0 | 1.0 | 1.0 | 5.238988 | 5.739315 | 8.507579 | 8.341367 | 10.823820 | 6.808361 |
| 4 | 0.0 | 0.0 | 0.0 | 0.334202 | 0.772579 | 2.889912 | 3.521248 | 4.585416 | 3.336505 |
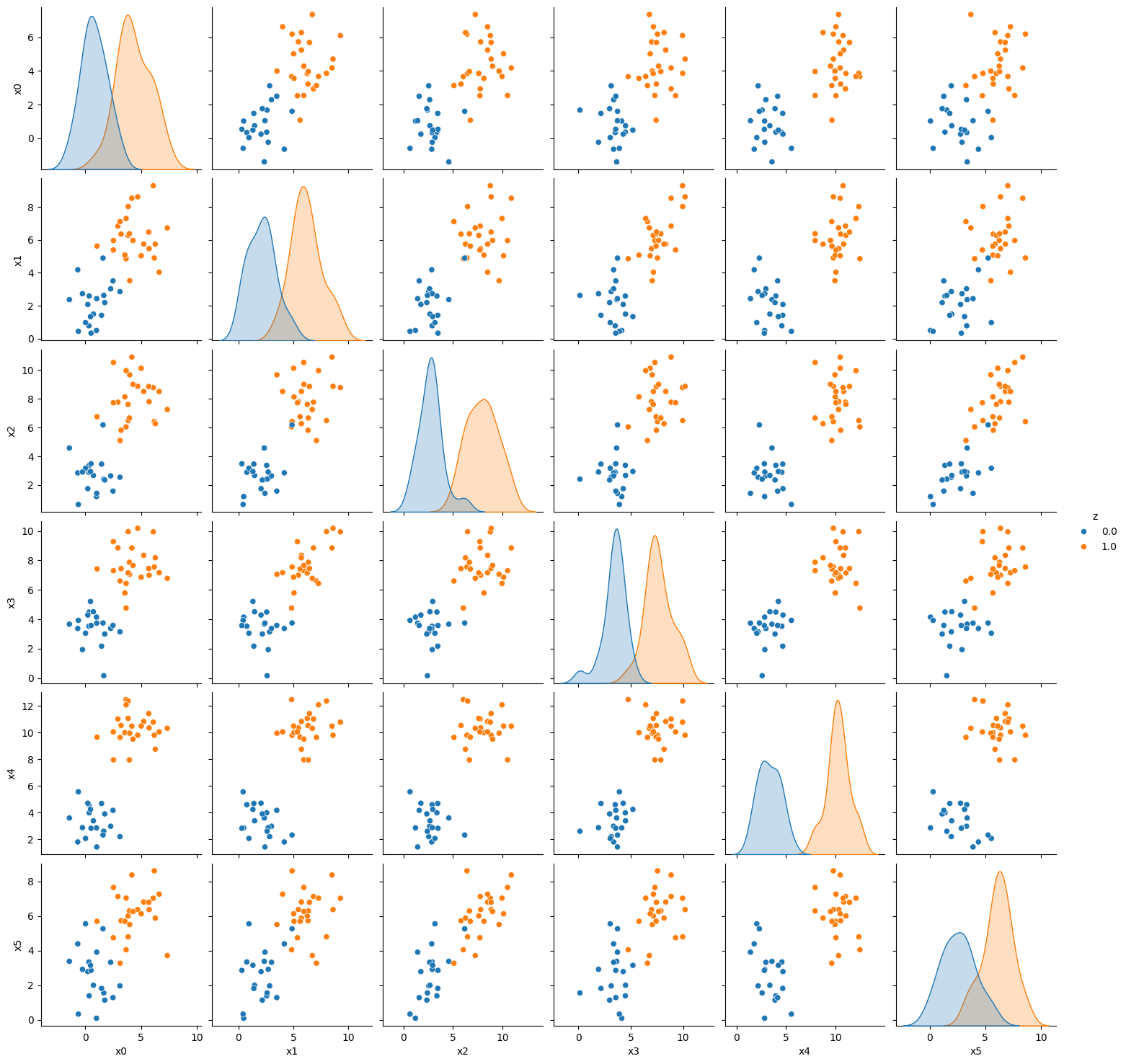
var_list = ['x'+ str(i) for i in range(D)]
g = sns.pairplot(pop_feature_bias_df1, vars= var_list, hue = 'z')
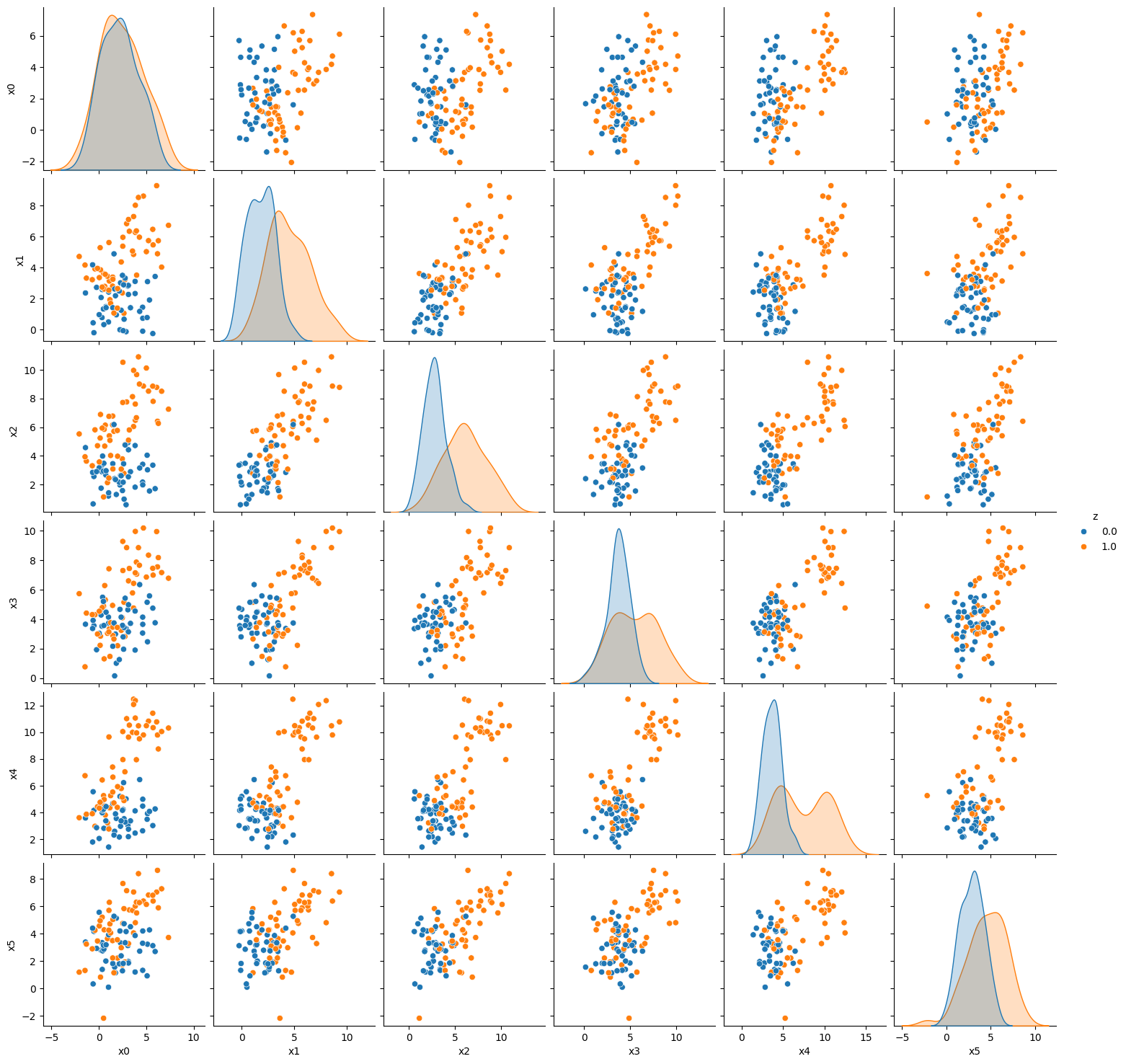
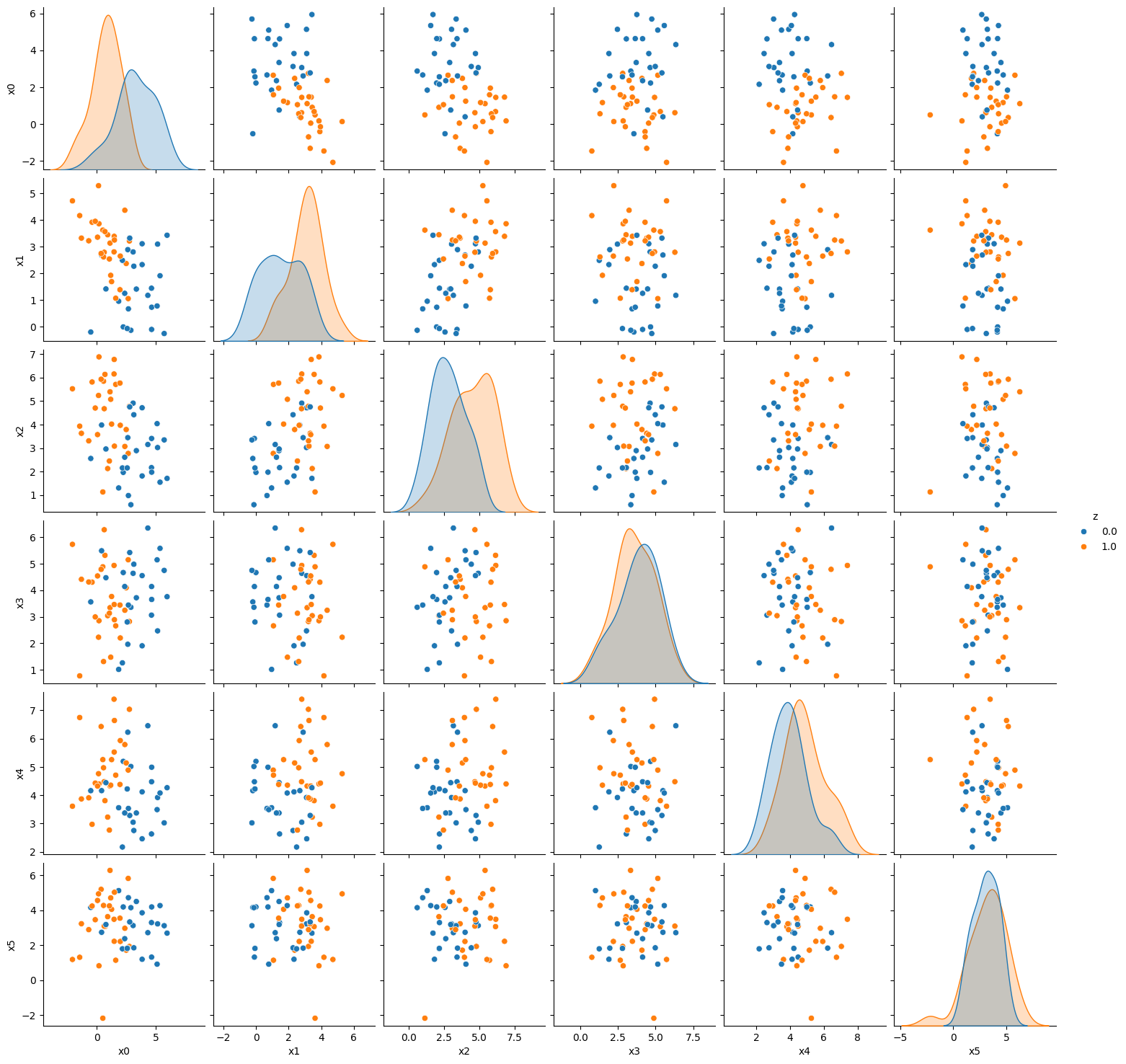
[sns.pairplot(dffbai, vars= var_list, hue = 'z') for ai,dffbai in pop_feature_bias_df1.groupby('a')]
[<seaborn.axisgrid.PairGrid at 0x7f3672059940>,
<seaborn.axisgrid.PairGrid at 0x7f367096a390>]